Fusion Hub Data Quick-Start#
This notebook demonstrates the process of:
Searching a collection to intersect a point location between a start data and end date
Converting the search result to an
xarrayDataArraySubsetting the DataArray to a geometry
Plotting the time series in a grid
Extracting data from a point location
Reducing the data within a geometry
Downloading a file based on valid data percentage
Downloading multiple files
Removing data entries based on some condition
Creating matplotlib animations
import json
import requests
import pystac
from pystac_client import Client
from pprint import pprint
from botocore.exceptions import ClientError
from shapely.geometry import box, mapping, Point, Polygon
from pyproj.crs import CRS
import geopandas as gpd
import xarray as xr
import rioxarray as rxr
import rasterio as rio
from matplotlib import pyplot as plt
import numpy as np
import os
import base64
If you see ERROR 1: PROJ: proj_create_from_database: Open of /opt/conda/share/proj failed, do not fret, and proceed.
This next cell opens a file creds.json which you will need to create in the same directory as the notebook. The format of the file should be:
{
"username":"your_username",
"password":"your_password"
}
and you have updated with your username and password.
with open('creds.json') as f:
creds = json.loads(f.read())
This next cell will endecode the username:password combination and use it to authorize access to the STAC API given by the cat_url endpoint.
userpass = f"{creds['username']}:{creds['password']}"
b64 = base64.b64encode(userpass.encode()).decode()
headers = {'Authorization':'Basic ' + b64}
cat_url = 'https://stac.hydrosat.com'
catalog = Client.open(cat_url, headers)
We’ll search for data in the starfm_predictions_modis_landsat and pydms_sharpened_landsat collections which intersect a point location between a start date and an end date and print out the number of items.
geom = {'type': 'Point', 'coordinates': [-120.21102905273436,36.535019209789]} # Point for Hanford, CA, USA
collections = ["starfm_predictions_modis_landsat", "pydms_sharpened_landsat"]
start_date = "2021-08-17T00:00:00Z"
end_date = "2021-10-30T00:00:00Z"
search = catalog.search(
collections = collections,
intersects = geom,
datetime = [start_date, end_date],
max_items = 500
)
# items = list(search.items()) # for pystac-client >= 0.4.0
items = list(search.get_all_items()) # for pystac-client < 0.4.0
items.reverse() # make the results ascending in time
len(items)
65
This next cell will determine if the data returned covers more than a single MGRS tile. If there is, choose one of the tiles to subset the list of returned items.
mgrs_tiles = []
for i in items:
for l in i.to_dict()['links']:
if 'element84' in l['href']:
mgrs_tiles.append(l['href'].split(r'/')[-1].split('_')[1])
print(f'number of tiles in query: {len(set(mgrs_tiles))}, {set(mgrs_tiles)}')
# if there is more than one tile, uncomment and execute this next line to choose the MGRS tile you are interested in
# items = [i for i in items if mgrs_tiles[0] in i.id]
number of tiles in query: 1, {'10SGF'}
Now we’ll pass the first 25 items to the FH_Hydrosat class and stack the items into an xarray DataArray. We’ll print out the DataArray to get a summary of its contents.
from FH_Hydrosat import FH_Hydrosat
res = FH_Hydrosat(items[:25])
stacked_res = res.stack()
stacked_res.ds.sortby('time')
<xarray.DataArray (time: 25, band: 1, y: 5490, x: 5490)>
dask.array<concatenate, shape=(25, 1, 5490, 5490), dtype=float32, chunksize=(1, 1, 2048, 2048), chunktype=numpy.ndarray>
Coordinates:
* band (band) int32 1
* x (x) float64 7e+05 7e+05 7e+05 ... 8.097e+05 8.097e+05 8.098e+05
* y (y) float64 4.1e+06 4.1e+06 4.1e+06 ... 3.99e+06 3.99e+06
spatial_ref int32 0
* time (time) object 2021-08-17T11:59:59.500000+00:00 ... 2021-09-1...
Attributes:
_FillValue: nan
scale_factor: 1.0
add_offset: 0.0ds = stacked_res.ds.sortby('time')
The DataArray is quite large if we try to access all of the data. For ease of computation, we’ll subset the DataArray by a polygon, which will be generated by creating a rectangular buffer around the point location by 1km on either side.
p_geom = Point(geom['coordinates'][0], geom['coordinates'][1])
point_df = gpd.GeoDataFrame({'geometry':[p_geom]}, crs=CRS.from_epsg(4326))
raster_crs = CRS.from_wkt(ds.spatial_ref.crs_wkt)
buffer_dist = 1000 # 1km in local UTM zone
poly_df = point_df.to_crs(raster_crs).buffer(buffer_dist, cap_style = 3) # square buffer
Let’s plot the polygon on a Folium map so we can see where we are extracting the data
import folium
# Use WGS 84 (epsg:4326) as the geographic coordinate system
df = gpd.GeoDataFrame(poly_df.to_crs(epsg=4326))
m = folium.Map(location=[p_geom.y, p_geom.x], zoom_start=13, tiles='CartoDB positron')
# add the polygon and centroid
for _, r in df.iterrows():
# Without simplifying the representation of each polygon,
# the map might not be displayed
#sim_geo = gpd.GeoSeries(r['geometry']).simplify(tolerance=0.001)
sim_geo = r[0].simplify(tolerance=0.001)
geo_j = gpd.GeoSeries(sim_geo).to_json()
geo_j = folium.GeoJson(data=geo_j,
style_function=lambda x: {'fillColor': 'orange'})
geo_j.add_to(m)
lat = sim_geo.centroid.y
lon = sim_geo.centroid.x
folium.Marker(location=[lat, lon]).add_to(m)
m
Now let’s clip the dataset with the geometry using rioxarray’s rio utility package
from FH_Hydrosat import FH_StackedDataset
# clip the raster dataset and cast to a class with slightly more functions
clipped = FH_StackedDataset(ds.rio.clip(poly_df.geometry))
ds_clip = clipped.ds
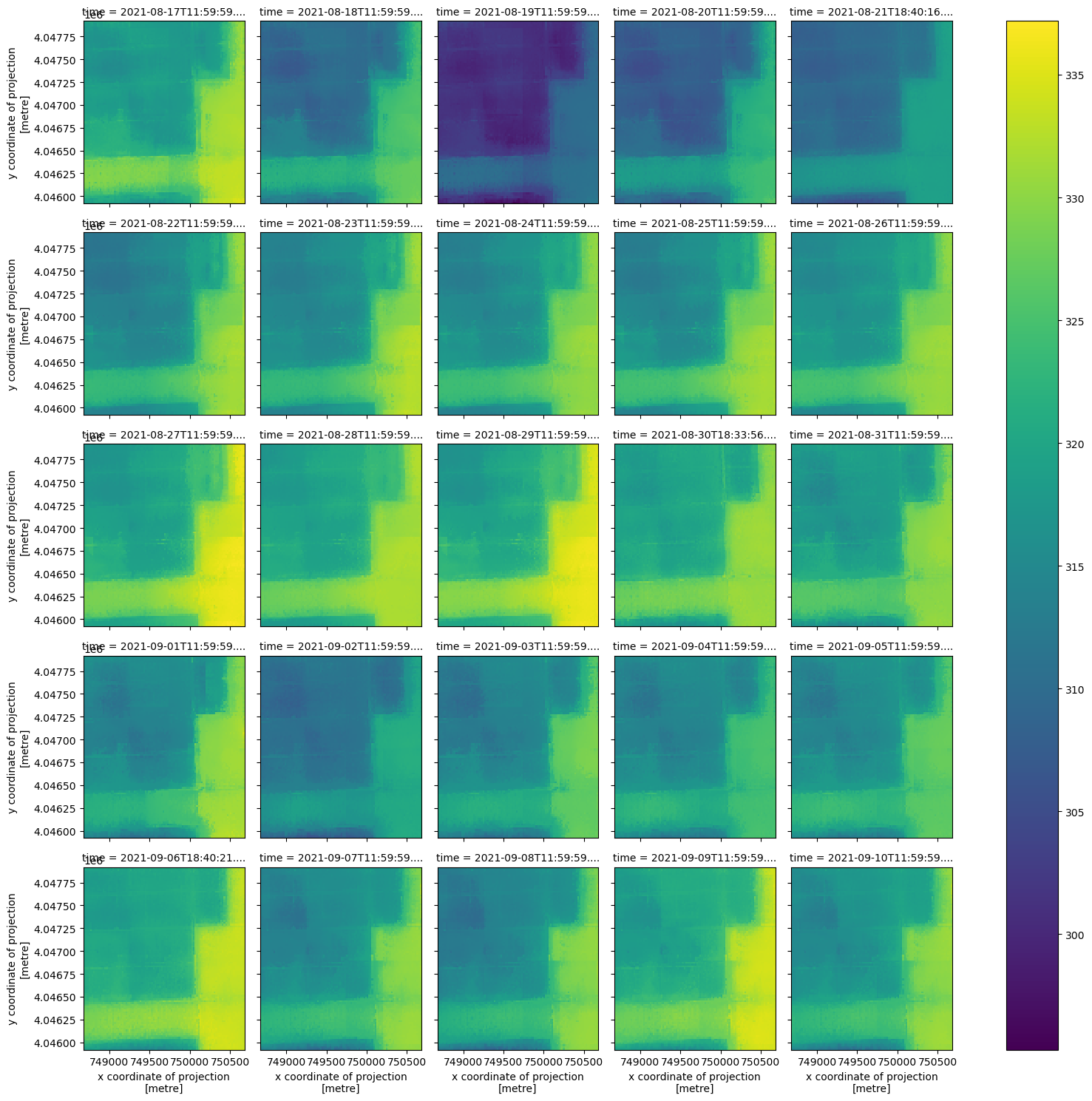
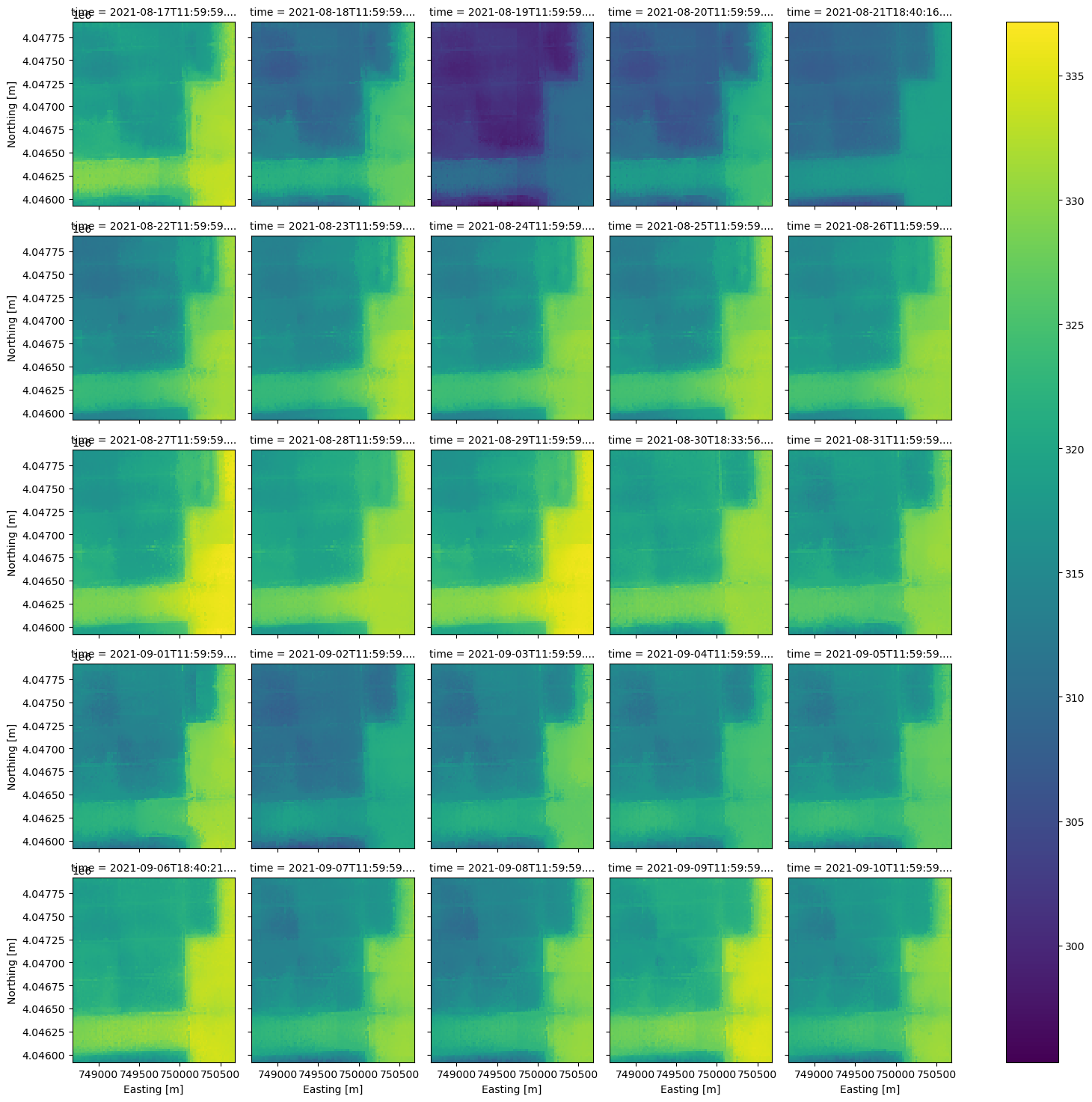
Now that we have a smaller DataArray, let’s plot the contents according to the time dimension using xarray’s plot utility.
ds_clip.plot(x='x', y='y', col='time', col_wrap=5);
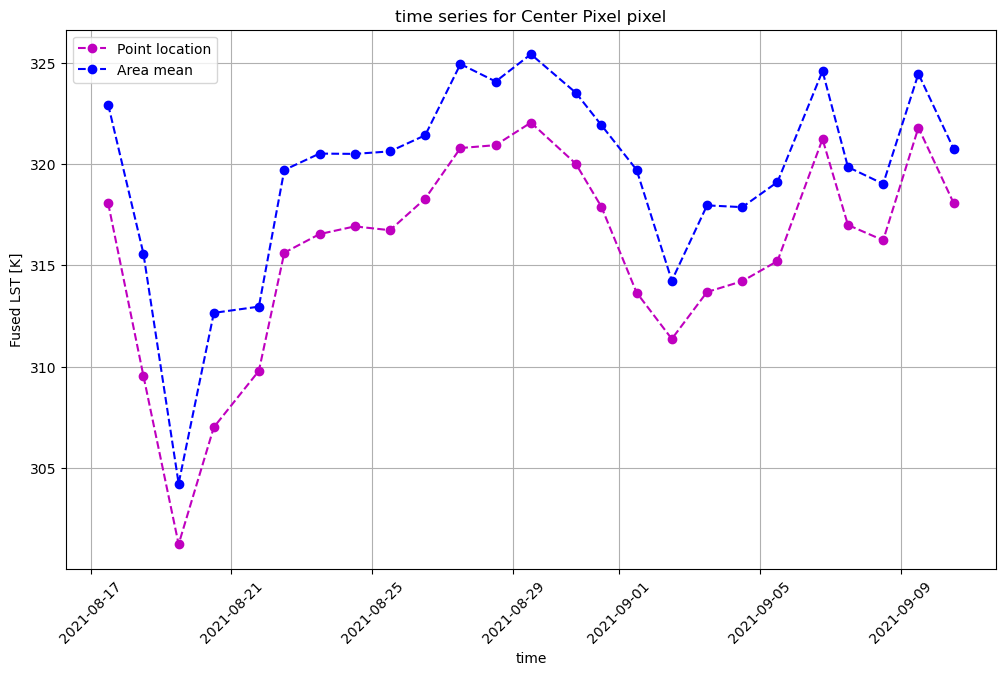
Plot a time series for the point location#
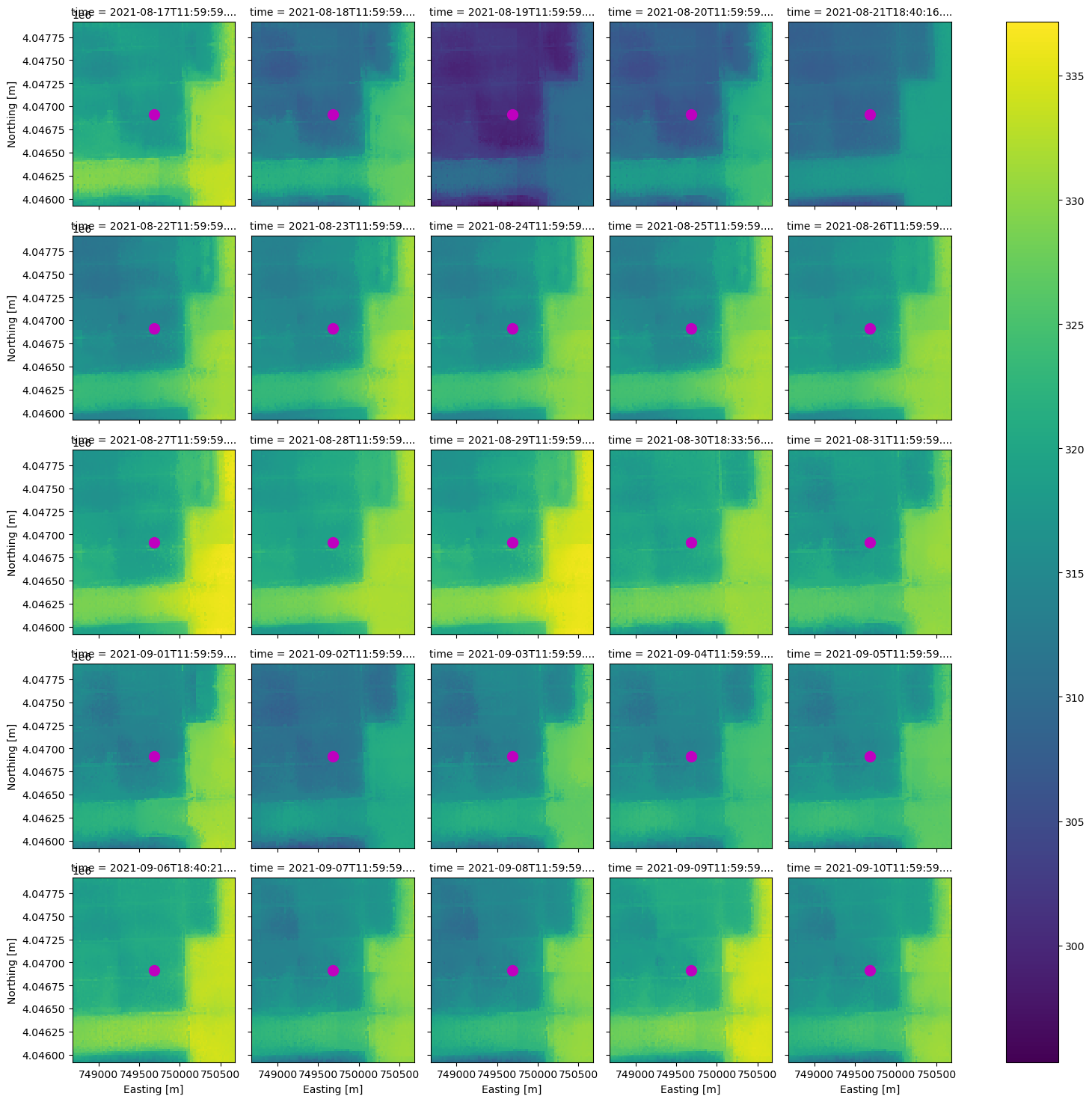
With the same DataArray, we can extract the pixel values which intersect a point location. Let’s use the same point location we used to search the STAC catalog and plot it.
centroid = poly_df.geometry[0].centroid
set_x, set_y, pixtype = (centroid.x, centroid.y, 'Center Pixel') # change 'Center Pixel' for plot title
ax = ds_clip.plot(x='x', y='y', col='time', col_wrap=5)
ax.set_xlabels('Easting [m]')
ax.set_ylabels('Northing [m]')
ax.map(lambda: plt.plot(set_x, set_y, markersize=20, marker=".", color="m"))
plt.show()
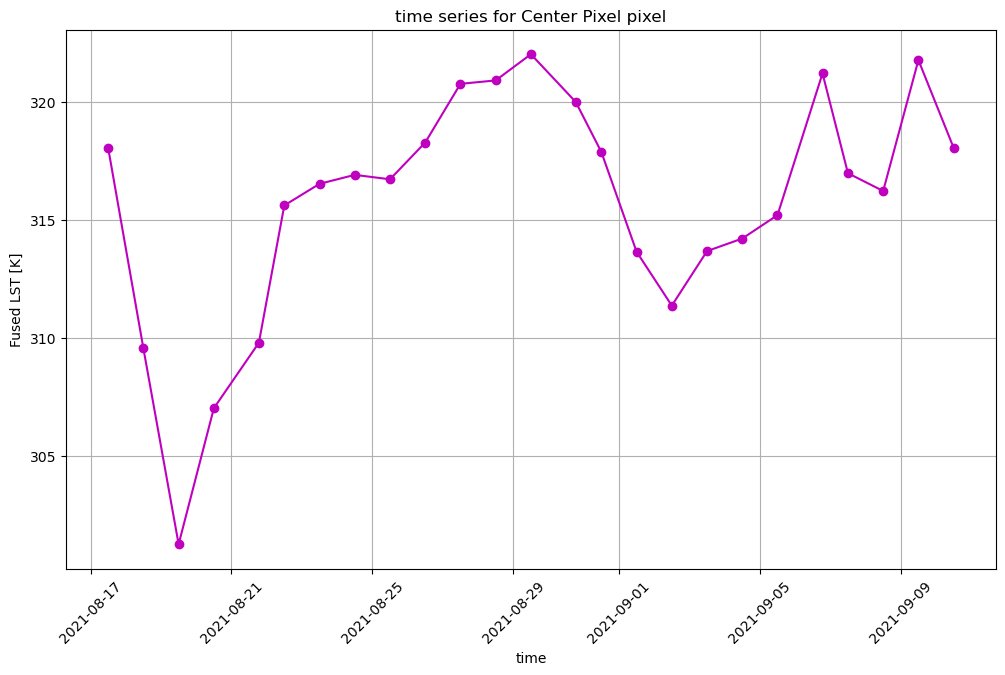
ax = ds_clip.isel(band=0).sel(x=set_x, y=set_y, method='nearest', tolerance=20).plot(marker='o', c='m', figsize=(12,7))
plt.title(f'time series for {pixtype} pixel')
plt.grid(True)
plt.ylabel('Fused LST [K]')
plt.xticks(rotation=45)
plt.show()
C:\software\Anaconda3\envs\hydrosat\lib\site-packages\xarray\core\indexes.py:234: FutureWarning: Passing method to Float64Index.get_loc is deprecated and will raise in a future version. Use index.get_indexer([item], method=...) instead.
indexer = self.index.get_loc(
C:\software\Anaconda3\envs\hydrosat\lib\site-packages\xarray\core\indexes.py:234: FutureWarning: Passing method to Float64Index.get_loc is deprecated and will raise in a future version. Use index.get_indexer([item], method=...) instead.
indexer = self.index.get_loc(
Similarly, we can reduce the area within the subsetting geometry to the mean value, and plot against the time series for the point location.
ax = ds_clip.plot(x='x', y='y', col='time', col_wrap=5)
ax.set_xlabels('Easting [m]')
ax.set_ylabels('Northing [m]')
ax.map(lambda: plt.plot(set_x, set_y, markersize=20, marker=".", color="m"))
plt.show()
ax = ds_clip.isel(band=0).sel(x=set_x, y=set_y, method='nearest', tolerance=20).plot(linestyle='--', marker='o', c='m', label='Point location', figsize=(12,7))
ax = ds_clip.isel(band=0).mean(dim=('x', 'y')).plot(linestyle='--', marker='o', c='b', label='Area mean')
plt.title(f'time series for {pixtype} pixel')
plt.legend()
plt.grid(True)
plt.ylabel('Fused LST [K]')
plt.xticks(rotation=45)
plt.show()

C:\software\Anaconda3\envs\hydrosat\lib\site-packages\xarray\core\indexes.py:234: FutureWarning: Passing method to Float64Index.get_loc is deprecated and will raise in a future version. Use index.get_indexer([item], method=...) instead.
indexer = self.index.get_loc(
C:\software\Anaconda3\envs\hydrosat\lib\site-packages\xarray\core\indexes.py:234: FutureWarning: Passing method to Float64Index.get_loc is deprecated and will raise in a future version. Use index.get_indexer([item], method=...) instead.
indexer = self.index.get_loc(
download a single file#
# find which file has the most data in it
max_data_idx = np.argmax(ds_clip.count(dim=('x', 'y', 'band')).values)
outfile = f'./{os.path.basename(res.item_desc[max_data_idx])}'
print(outfile, os.path.exists(outfile))
res.download_single_asset(max_data_idx)
print(outfile, os.path.exists(outfile))
./10SGF_pred_20210817_from_20210814_using_MODIS_Landsat.tif True
./10SGF_pred_20210817_from_20210814_using_MODIS_Landsat.tif True
download multiple files, since there a few scenes that have full coverage for the AOI#
# download the items above which correspond to items 2,5,6, and 10
# using the download_multiple_assets() function
res.download_multiple_assets([2,5,6,10])
using 2 processes to download 4 assets
make an animation from the full set of data#
If you see Javascript Error: IPython is not defined or MovieWriter ffmpeg unavailable; using Pillow instead, do not fret, and proceed.
from IPython.display import HTML
# switch plotting to allow for animations
%matplotlib notebook
# use the create_animation() function.
# display the animation with HTML display
ani = clipped.create_animation(save_ani=True, vmin=300, vmax=350)
HTML(ani.to_jshtml())
drop files with data less than 50% coverage#
Sometimes the data will have low coverage due to cloud cover or invalid pixels in the input data products. Let’s remove them from the dataset using the remove_below_data_perc() function, plot them, and create an animation.
# to switch back and allow xarray plots
%matplotlib inline
more_data = FH_StackedDataset(clipped.remove_below_data_perc(clipped.ds, 0.5))
ax = more_data.ds.plot(x='x', y='y', col='time', col_wrap=5)
ax.set_xlabels('Easting [m]')
ax.set_ylabels('Northing [m]')
plt.show()
# switch back to allow animation
%matplotlib notebook
ani = more_data.create_animation(save_ani=True, vmin=300, vmax=350, anipath='more_data.gif')
HTML(ani.to_jshtml())
You may notice that the animation plays with the most recent date in the data stack first, and then ascending. To reverse the order to descending from the last date in the dataset, use an xarray call to reverse the time dimension on the dataset.
# reverse the order
more_data_rev = FH_StackedDataset(more_data.ds.sortby('time', ascending=False))
ani = more_data_rev.create_animation(save_ani=True, vmin=300, vmax=350, anipath='more_data_rev.gif')
HTML(ani.to_jshtml())